Index
| Table of Contents | ||||
|---|---|---|---|---|
|
General
For this exercise, you need either Pymol or Yasara installed that has the 3DM plugin. If you don't have Yasara or Pymol or you are missing the 3DM functionality, please consult the installation instructions. Before you start this exercise make sure you have the latest version of Yasara or Pymol installed.
Login at 3DM with your 3DM account. If you don't have a 3DM account you can request one via the "get 3DM" tab. To be able to do this course you need to have access to the databases used in this course. After you have requested an account you can request access to the course databases by sending an email to joosten@bio-prodict.nl.
After entering the login details you will see a 'Select 3DM system' page. Click on the 'Public 3DM Systems' checkbox and search for the "Phosphoenolpyruvate mutase/Isocitrate lyase" database. Open this 3DM system.
...
| Panel | ||||
|---|---|---|---|---|
| ||||
At the starting page of each 3DM database, you see the 3DM data cycle. The icons in the circle represent links to the most important 3DM options. These options are also available on the left. |
Introduction
Fungi can be pathogenic to plants and animals. It is known that the secretion of oxalate by fungi is a commonly used strategy for their pathogenicity. Oxalate is toxic and can form crystals that demolish the cell wall of the host. The oxalate is produced from oxaloacetate catalyzed by the enzyme oxaloacetate hydrolase (OAH). This is the reaction:
...
On the protein information pages, you can find a couple of different tabs. Have a quick look at what you can find in each tab.
Subsets
3DM offers several ways to select a subset of sequences. Once a subset is selected a mini 3DM can be generated for this subset. All 3DM functionalities, such as the correlated mutations, are regenerated and can separately be analyzed. The data of a subset can also be compared to the data of the full set of sequences or with other previously defined subsets.
...
| Expand | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||
Position 157 scores highest when both numbers are added. Positions that both make a contact with a ligand but also show correlated mutation behavior are likely hotspots for specificity.
Some positions, like 116 make a lot of contacts with ligands but do not show correlated mutation behavior. Do you understand why this is? Position 116 is a conserved position. This position is important for the general function of the protein (the reaction) and not for the specific function.
|
Homology modeling of OAH niger with its substrate oxaloacetate and the design of an inhibitor
Build a homology model
- Go back to the protein information page of G3Y473 and select the ‘MODELS' tab.
...
| Info | ||||
|---|---|---|---|---|
| ||||
What is the residue type of 157? In yasara you can make a residue visible by right-clicking on the residue in the sequence at the bottom of Yasara and choose "Show Atoms → residue" |
| Expand | ||
|---|---|---|
| ||
Serine. |
Load an inhibitor
Structures can be loaded directly in Yasara from the 3DM database via the 3DM → Structures → load structure from 3DM option. Loading structure files via the 3DM menu ensures that the structures are all superimposed, co-crystallized compounds will have to be positioned in the active site and proteins will have the 3D numbering.
- Load the inhibitor of 1M1BA using the "load data from 3DM" option in Yasara.
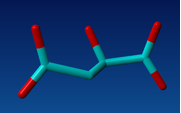
Build oxaloacetate from this inhibitor.
Fig 2. Structure of oxaloacetate.
...
Fig 6. Picture of the model of OAH taken from the 2008 publication: Identification of fungal oxaloacetate hydrolyase within the isocitrate lyase/PEP mutase enzyme superfamily using a sequence marker-based method. This picture clearly shows the predicted Ser157 H-bridge with the diol of oxaloacetate.
Extra questions
Position 157 is the center of the correlated mutation network. P is the most common residue at position 157 (is that correct?). We have generated a subset of sequences that have a P at position 157 called "P157"
...