...
To view 3DM data inside protein structures and models, use these guides to install the 3DM plugin for your molecular viewer of choice:
Test the Yasara 3DM connection
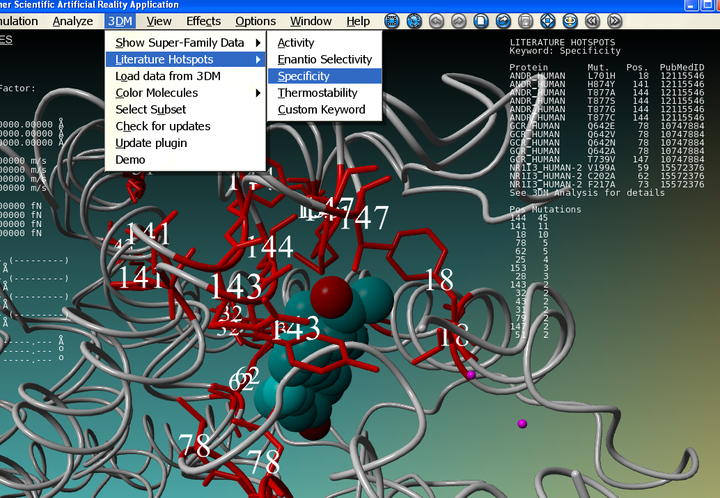
If you have requested the free Yasara View the 3DM-Yasara connection only works for the nuclear receptor ligand binding domain structures. You can try out how it works using nuclear receptor structures. For convenience we have prepared an example scene file containing two nuclear receptor structures which can be downloaded here and opened with Yasara (File -> Load -> Yasara Scene). Just try out the different 3DM options and see what happens. If asked for a password, use your Bio-Prodict Account credentials. You can also find 3DM data by right clicking on amino acids. Note that the data in the Heads Up Display is clickable to retrieve the underlying data (figure 4).
By selection structures from the 3DM system yasara will understand the 3D alignment numbering used in the 3DM alignment. This ensures the coupling of the structures to other data in the 3DM database. There are two ways to select structures from 3DM:
- Use the 3DM option in Yasara -> “select data from 3DM” and give a PDB name (e.g. 1A28).
- You can also select structures directly from the 3DM websites (first login via 3dm.bio-prodict.com then select the nuclear receptors – ligand binding domain demo 2008 system or go to your own 3DM if you have one). Once logged in you can export structures to Yasara by generating a Yasara scene via the “visualize data in structures” option once you have logged into the demo nuclear receptor 3DM system. This is how the example scene file was generated.
...
Read about 3DM, or do the Course
...