For this exercise, you need either Pymol or Yasara installed that has the 3DM plugin. If you don't have Yasara or Pymol or you are missing the 3DM functionality, please consult the installation instructions. Before you start this exercise make sure you have the latest version of Yasara or Pymol installed.
Login at 3DM with your 3DM account. If you don't have a 3DM account you can request one via the "get 3DM" tab.
After entering the login details you will see a 'Select 3DM system' page. Click on the 'Public 3DM Systems' checkbox and search for the "Phosphoenolpyruvate mutase/Isocitrate lyase" database. Open this 3DM system.
At the starting page of each 3DM database, you see the 3DM data cycle. The icons in the circle represent links to the most important 3DM options. These options are also available on the left. |
Fungi can be pathogenic to plants and animals. It is known that the secretion of oxalate by fungi is a commonly used strategy for their pathogenicity. Oxalate is toxic and can form crystals that demolish the cell wall of the host. The oxalate is produced from oxaloacetate catalyzed by the enzyme oxaloacetate hydrolase (OAH). This is the reaction:
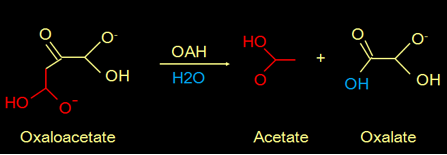
Fig 1. Reaction mechanism that produces oxalate.
We have generated a 3DM for the corresponding protein family. OAH falls in the Phosphoenolpyruvate mutase/Isocitrate lyase superfamily.
The OAH of niger is the best characterized OAH protein. This is the sequence:
>G3Y473 MKVDTPDSASTISMTNTITITVEQDGIYEINGARQEPVVNLNMVTGASKLRKQLRETNEL LVCPGVYDGLSARIAINLGFKGMYMTGAGTTASRLGMADLGLAHIYDMKTNAEMIANLDP YGPPLIADMDTGYGGPLMVARSVQQYIQAGVAGFHIEDQIQNKRCGHLAGKRVVTMDEYL TRIRAAKLTKDRLRSDIVLIARTDALQQHGYDECIRRLKAARDLGADVGLLEGFTSKEMA RRCVQDLAPWPLLLNMVENGAGPVISVDEAREMGFRIMIFSFACITPAYMGITAALERLK KDGVVGLPEGMGPKKLFEVCGLMDSVRVDTEAGGDGFANGV |
For each protein in the 3DM database, there is a "protein information" page that contains more detailed information.
Find the protein information page of the sequence above using the search option of 3DM. |
| In the quick search (you can find box this just above the green bar in 3DM) you can use "G3Y473” or you can simply search for G3Y473 in the keyword search tab. |
On the protein information pages, you can find a couple of different tabs. Have a quick look at what you can find in each tab.
3DM offers several ways to select a subset of sequences. Once a subset is selected a mini 3DM can be generated for this subset. All 3DM functionalities, such as the correlated mutations, are regenerated and can separately be analyzed. The data of a subset can also be compared to the data of the full set of sequences or with other previously defined subsets.
With the search option we have made a subset called "oxalate producers" that contains the proteins available in this 3DM system for fungi of which it is known that they can produce oxalate:
Aspergillus clavatus Neosartorya fischeri Penicillium chrysogenum Penicillium marneffei Talaromyces stipitatus Sclerotinia sclerotiorum Aspergillus niger Sclerotium cepivorum Aspergillus terreus Aspergillus fumigatus Botryotinia fuckeliana |
Do you understand how we generated this subset (e.g. which search options we combined in the subset window → don't actually do this yourself, guest accounts cannot make subsets)? |
In the keyword search tab of the search option, you can select species. Here you can type the species names. We have separately searched for each of the species and the resulting proteins were added to the subset window by clicking the + signs of the subset window. Try to search yourself for Aspergillus clavatus in the species search options. You will find two proteins (A1CFP3 and A1CMM8). These are the first two proteins of the subset. You can get a list of the proteins that are in a subset by clicking in the subset window on the number (in this case 33) that indicates how many proteins are in the subset. |
Do you understand why we generated this subset? What do you think we can learn from this subset? |
A subset is a mini 3DM made of only the sequences that are defined in the subset. All features and data types are re-calculated and can be compared to data in other subsets of to the whole superfamily. Here we want to find the proteins/residues responsible for oxalate production in oxalate-producing fungi. We try to find things (e.g. residues) that are specific to the subset. |
At the alignment, statistics pages change between the full dataset and the "oxalate producers" you just made using the 'Subset' menu in the header on top of 3DM and see how the graphs change.
3DM always generates an extra histogram for each subset that shows the residues that are specifically conserved in the selected subset (the histogram called "subset specific conserved residues"). The highest scoring residues are around 3D positions 157.
Important here is to realize that these are positions that are not just simply conserved in this subset of oxalate producing fungi, but the corresponding residues are absent from the rest of the sequences in the superfamily. In other words, these residues are specific for this subset.
You can see this by comparing this plot with the amino acid conservation plot of the new subset. Use the "custom plot" tab and select your subset from the left box and from the right box "amino acid conservation " and "subset specific conserved residues".
How many positions are 100% conserved? And how many of those are specific for the oxalate-producing fungi? |
You can put the slider bar of the amino acid conservation plot on 100%. You will see that there are 47 positions 100% conserved in the oxalate producers subset (see figure).
This figure shows what you should have selected. Here the conservation cut-off was set at 100%. Clearly, the subset-specific conserved residues are found mainly around position 157. |
On the alignment page:
What is the most conserved residue at this position in the "oxalate producers" subset? |
S157 → 100% in the subset. |
And what is the percentage of this residue in the full alignment (hint: Use the subset menu in the header to change between your subset and the full dataset). |
S157 → 1.06% in the full alignment. |
So what is the difference? Do you understand the "subset specific conserved" plot from the previous question? |
Difference = 98.94%. In the "subset specific conserved" plot the number is 99,38 (you can see this by putting your mouse over the peak of the subset specific conserved plot at position 157). The difference between these numbers comes from the fact that in the full alignment the serine's of the oxalate producers subset are included. The "subset specific conserved residues" plot calculates the difference between the conservation in the subset minus the conservation of the full set but then without the sequences of the subset. |
Take home message: the data you are looking at is always depending on the subset tab that is selected. |
Click on "Correlated mutations" in the menu on the left. Make sure you have the "Full Dataset" selected at the top of 3DM.
Correlated mutations calculated for a superfamily alignment often reflect positions important for specificity because superfamily alignments contain enzymes with different specificities (do you understand this concept?)
Explain this concept. |
The proteins in a superfamily usually form groups of different specificities. Within a group the residues important for specificity are conserved, but between the groups, they mutate. Since they all mutate simultaneously between the groups they result in a correlated mutation network. Note: which protein feature is behind a correlated mutation network heavily depends on the input alignment. If you make a subset of enzymes that all have the same specificity the correlated mutation will, of course, not reflect changes in specificity and thus the network will not be composed of specificity hotspots. The concept of how to choose the input alignment is explained in more detail later in this practical. |
The "Top Correlation Heatmap" page shows the alignment positions of which the residues mutate simultaneously (definition of a correlated mutation).
Which position is the highest correlating? |
Position 157 is the highest correlating position. Note that you can click on the heatmap. This will lead to plots showing the amino acid distribution of the two corresponding positions. Those plots show the co-occurrence of amino acids. This data can be use to see if it might be better to make certain double mutants instead of single mutants. |
Do you see why it is so handy to have the 3D numbers in the network? |
It enables to the plot of any other data type of which the 3D number is known on the network. This is one of the strong features of 3DM. All data and all tools are connected via 3D numbers. |
Which residue positions are reported to affect specificity and which one is the most published position in relation to specificity? |
Giving the keyword specificity in the search box results in 5 papers of position 157. |
3DM calculates an enrichment score for a given keyword. What do you think this enrichment score means? |
The enrichment score is the factor that shows how many more times mutations related to the keyword are found in the network compared to positions outside the network. The enrichment score for specificity in the OAH network is 7.39. This means that there are 7.39 times more mutations affecting specificity published at positions in the network. Thus it is likely that specificity is the evolutionary pressure that caused the positions in the network to mutate simultaneously. Note that an enrichment score of >4 or 5 normally is significant. |
If you would add these plots what is the highest scoring position? What does this mean? |
Position 157 scores highest when both numbers are added. Positions that both make a contact with a ligand but also show correlated mutation behavior are likely hotspots for specificity.
Some positions, like 116 make a lot of contacts with ligands but do not show correlated mutation behavior. Do you understand why this is? Position 116 is a conserved position. This position is important for the general function of the protein (the reaction) and not for the specific function.
|
3DM selected three structures as potential good templates. In this course, you will learn how to select the best template, make the best alignments, etc., but for now, we will use 3LYEA → it does have the best resolution (e.g. quality).
Select 3LYEA as a template and use the "alignment" as numbering. You can open the model either with Yasara or Pymol. Note that generated models can always be retrieved from the "visualize" pages. The third form on this page contains the models. Here you can select your model and different data that you want to visualize in your model.
What is the residue type of 157? In yasara you can make a residue visible by right-clicking on the residue in the sequence at the bottom of Yasara and choose "Show Atoms → residue" |
Serine. |
Structures can be loaded directly in Yasara from the 3DM database via the 3DM → Structures → load structure from 3DM option. Loading structure files via the 3DM menu ensures that the structures are all superimposed, co-crystallized compounds will have to be positioned in the active site and proteins will have the 3D numbering.
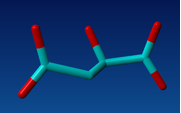
Fig 2. Structure of oxaloacetate.
This is the structure of oxaloacetate. We are very lucky since it is very similar to the structure of the 1M1BA inhibitor. Simply swapping the SO3 group with a CO2 group will do the job.
In Yasara:
In Pymol:
The reaction mechanism of isocitrate lyase (ICL) is known for quite a while (fig 3). In this reaction mechanism the H of the blue OH group donates an electron, makes a double bond, and splits of the COOH group.
Do you think OAH can use the same reaction mechanism to break down oxaloacetate? |
No, it has a =O instead of an OH. |
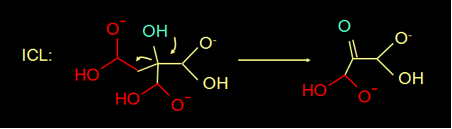
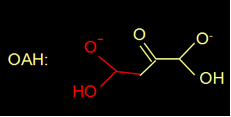
Fig 3. Reaction mechanism of ICL (above) and the structure of oxaloacetate (below)
Actually, oxaloacetate in water is in equilibrium with its diol form (figure 4).
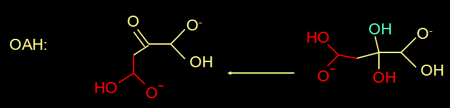
Fig 4. Oxaloacetate is in equilibrium with its diol.
Do you think this diol of oxaloacetate can be converted with the same reaction mechanism as ICL? |
Yes the diol form of OAH has the required OH group. |
Until today OAH is the only known enzyme of this superfamily that has a substrate in a diol form. So the extra OH is unique to OAH.
Where do you think the extra OH will be positioned? |
The OH unique to OAH is sticking right towards ser157. |
Can you think of a reason why the Ser157 is also unique to OAH? |
Because oxaloacetate is the only substrate that has this diol form the Ser contacting this extra OH is unique to OAH. |
Modeling the extra OH in the active site with the "swap" option does not work very well in yasara, because yasara can't deal with changing the double bond of C=O to the single bond of C-OH without proper energy minimization (try to make the diol with the swap option if you like).
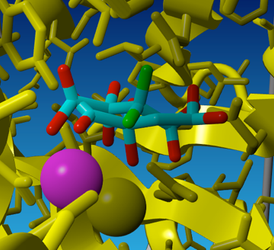
Fig 5. The result of energy minimization performed on the diol form of oxaloacetate in the OAH model.
In 2008 a model of OAH was generated similar to the way you did it today. With this model we were already in 2008 able to:
The inhibitor was designed by organic chemists that realized they had to make a compound that is 100% in the diol form. This was the case with difluoro-oxaloaceate. This compound indeed proved to be a very strong inhibitor of OAH and was later crystallized together with OAH of the fungus Cryphonectria Parasitica (pdb file 3M0JA).
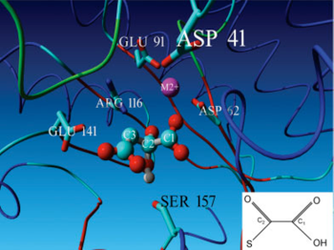
Fig 6. Picture of the model of OAH taken from the 2008 publication: Identification of fungal oxaloacetate hydrolyase within the isocitrate lyase/PEP mutase enzyme superfamily using a sequence marker-based method. This picture clearly shows the predicted Ser157 H-bridge with the diol of oxaloacetate.
Position 157 is the center of the correlated mutation network. P is the most common residue at position 157 (is that correct?). We have generated a subset of sequences that have a P at position 157 called "P157"
Do you think 157 will show a high correlated mutation score in this subset? |
No, in a subset with only P on position 157 the P is, of course, 100% conserved and therefore can't mutate together with other positions. The correlated mutation data that 3DM calculates is a measure of how often mutations occur together between two positions. No mutations will result in a score of 0. |
Using Yasara or Pymol, investigate the new correlated mutation network. Can you find the role of the amino acids in this new network (what is the function behind this network)? |
In the figure, you can see where the correlated mutation is found in the P157 alignment. This plot was generated by clicking "visualize all notes" in CorNet when the P157 subset was selected.
From within Yasara you can also make this visualization by the following functions:
Now to answer the question do the following:
You can switch between the two scenes using the different tabs. Clearly, the two scenes overlap. So the correlated mutations in the P157 alignment are formed during evolution in the dimerization domain.
|
Take home message: The function underlying correlated mutations heavily depend on the input alignment. Always look for additional data (in this case protein-protein interaction data → did you find that?) that might explain a correlated mutation network. |
The correlated mutations in this superfamily seem to reflect positions important for specificity. You want to change the specificity of OAH and you decide to rationally design a mutant library. Your screening method allows you to screen up to 1000 mutant clones.
How would you design your library? (Just give a general description of which residue positions you would choose, why you choose those and which residues you would try at those positions) |
This question can be answered thoroughly or just very simple. One thing is for sure, everything shows that the correlated mutations are important for specificity. Those are your first-choice hotspots. Then pick as many positions as your screen allows only using common residues at these hotspots (e.g. residues > 2% or so). This cut-off percentage is depending on how many hotspots you want to use. The more hotpots the higher this percentage or your library size gets too big. So it is always a trade-off between the number of hotspots and the number of residues per hotspot. There are many things to consider when you choose hotspots and the residues at the hotspots. Each position should be considered carefully and all data at each position should be investigated. These are things to consider:
|