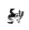...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
...
Intro
Welcome to the 3DM tutorial. A 3DM system is a data integration platform, we collect and integrate data about a protein superfamily.
...
You have access to more than 24k systems, each containing from 2k to 300k sequences.
Protein detail page
For now we're going to work on a UniProt protein E2R0G7 (soon the same functionalities will be available for connected Virus-X proteins)
...
Let's now again go back to the INFORMATION tab and click on the link in the aligned in subfamily field, show protein in subfamily alignment - the ID is the PDB id of the structure that was used as a template for the subfamily alignment.
Alignment page
Now we're at the alignment where your protein of interest is aligned. The displayed residues are only residues that are aligned in the core regions - core regions is part of the alignment that is aligned across the whole superfamily.
...
The lighter coloured residues are the variable regions and the bright-coloured ones are core regions. Keep in mind that only the aligned parts of the variable regions are displayed so you often don't see the full sequence in this view.
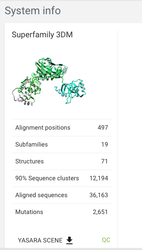
System info
Let's now have a look at the system info page - you can find a link in the menu on the left. This shows you an overview of the system, and gives you an idea of how much data there is.
Alignment statistics
Let's now navigate to the alignment statistics page, which you can access from the menu on the left side of the page.
...
Another thing you can do here is visualise this data in yasara - to do that click on the little button with a protein helix symbol . You will be redirected to the visualize page, but don't do it for now, we'll get to it later.
Alignment position page
You can also view data that's mapped onto a certain position in the alignment - the alignment position pages can be accessed in multiple ways, e.g.
...
On the histogram scroll to the right and click on the bar for position 214. Now we can take a look around the alignment position page. Here in the different tabs you can see what data is mapped onto this position across all proteins in the 3DM systems. For example, in the mutations tab you can see all the mutations that we've found in the literature for this alignment position across all proteins in the system.
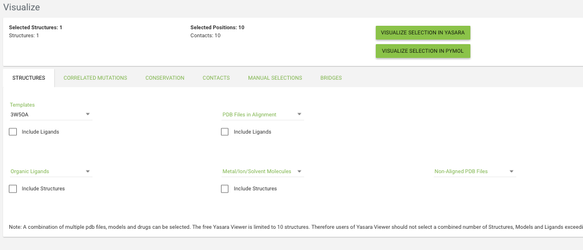
Visualize
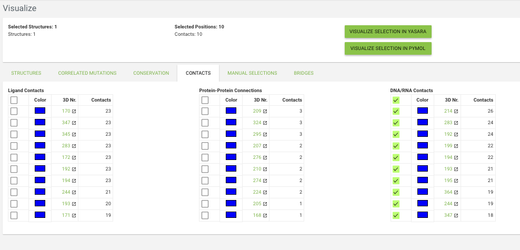
Go to the Visualize page from the menu, and click on the structures tab. From the templates menu select the 3W5OA structure - this is the template of the subfamily where our protein of interest is aligned. From the other tabs you can choose what data do you want to see mapped onto the structure - by default the residues with highest conservation and with highest correlated mutation score will be highlighted.
Go to the contacts tab and click on the checkbox in the top row in the DNA/RNA Contacts table (by clicking on the checkbox in the top row you toggle between selecting and deselecting all positions). Now click on the VISUALIZE IN YASARA button and a yasara scene with the selected data mapped onto the 3W5OA structure will be downloaded.
YASARA
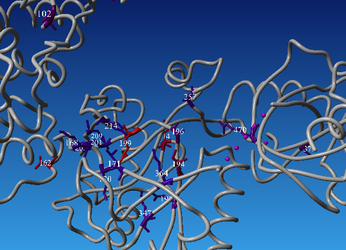
Open the downloaded scene in YASARA. You can see that some parts of the backbone are green(ish) and some are gray - the gray color indicates that these residues fall in the variable regions, while green are core residues.
...
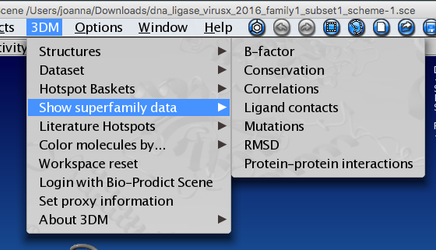
You can also access 3DM data from within yasara using our 3DM plugin - in the menu bar there is a '3DM' tab - there are numerous options of mapping your data onto the structure(s). Let's for example have a look at the mutation data. Go to 3DM > Show superfamily data > Mutations - now residues corresponding to alignment positions with the highest number of mutations are shown - you can see that they are mostly located in the pocket where you previously saw a lot of residues with DNA contacts. It makes sense that these residues are the ones that are most often mutated by researchers to investigate their function and the effect of mutations on DNA binding.
Phylogeny
Now click on the phylogeny item from the menu on the left. This shows an overall phylogeny tree where each node is one subfamily template. When you mouse over a subfamily ID you can display more information about the template structure.
Search options
There are multiple ways to search proteins in a 3DM system. While most of them are quite straightforward and probably don't need to be explained, we'll have a closer look at the 'Search proteins by position motif' and 'Search proteins by sequence motif'.
...
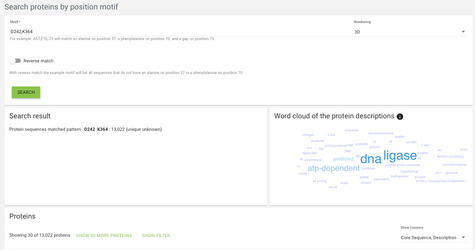
In the case of search proteins by position motif, we're looking for specific motifs on specific positions - for example we might want to find all proteins that have an aspartate on alignment position 242 and a lysine on alignment position 364
Virus-X proteins
Virus-X proteins are proteins that are added to the system through the Virus-X pipeline, they're only accessible for the Virus-X users.
...
For now there is just one protein, but later on we'll be adding more proteins from the Bielefeld's webserver.
Advanced
Numbering schemes
You can simplify your work with the protein of interest even more by creating a custom numbering scheme - that will cause the alignment positions to be renumbered to match the residue numbering of your protein.
...
To switch between the different numbering schemes click on the dropdown menu in the numbering scheme at the top of the page. And don't worry, creating new numbering schemes doesn't erase the previously existing ones, so after creating a custom numbering scheme you can still switch to the original 3DM numbering or other numbering schemes that you created.
Subsets
If we want to analyse only some of the proteins present in the system - for example only the closest homologs of the query protein than we'll need the subsets functionality. We're going to create a subset of 100 closest proteins to our E2R0G7 protein. To do that we need to again go to the protein detail page E2R0G7 and click in the sequence tab.
...
Note that you can also create subsets from results from all the other searches not only BLAST search - these will be described in the next section.
Hotspots
You can also use 3DM to find hotspots - important residues, affecting e.g. protein specificity or thermostability.
What if you don't have a protein of interest?
Panel design - protein selection tool
This is a tool to facilitate the design of enzyme selection panels (for example based on the sequence diversity on the selected hotspots). For this you're gonna need a more advanced course.
...
You can mail us or use the "Send feedback" form (linked on the bottom of the page)
Appendix
Correlation network and enrichment
Let's now go back to the website. Click on the correlated mutations item in the menu on the left. Now you see a network of correlated mutations - you can play around with the score cut-offs using the slider on the right. What you can also do is check if the positions involved in this network have any mutations with certain keywords assigned to them.
...